Research
Research Summary
The lab’s research focuses on using natural variation in genomes to understand evolutionary processes. A combination of modern molecular and computational approaches and the interpretation of physiological, evolutionary, and ecological contexts drive my research. In my research lab, we combine both historical and modern physiological data with new molecular genetic and genomic data to study the evolution of interactions between the environment and the genome, the evolutionary divergence of natural populations and sibling species, and the evolution of vector competency in malaria-carrying mosquitoes.
Using Next-Gen Sequencing to develop markers for phylogeographic analysis
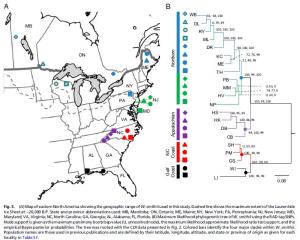
Studying the relatedness of recently diverged populations within species has been limited by the lack of genetic markers that can resolve phylogenetic patterns. In collaboration with members of the Cresko lab and the Bradshaw-Holzapfel lab, we have developed new methods using sequenced RAD-tags (sequenced on the Illumina sequencing platform) to simultaneously discover and type populations for thousands of genome-wide markers. Using markers that are fixed within and variable among populations, we then apply traditional phylogenetic methods to infer the relatedness of populations. Using the pitcher-plant mosquito, Wyeomyia smithii, we are able to show that all post-glacial populations have descended from a glacial refugium in the southern Appalachian Mountains of Eastern North America. The new RAD-tag method increased phylogenetic resolution several orders of magnitude over other genetic markers (mitochondrial genes and allozymes). [Figure taken from our recent PNAS paper here]
What is the genetic architecture underlying the evolution of photoperiodic time measurement (PTM)?
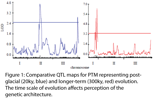 The evolution of PTM is a complex process, involving many currently unidentified genes. We performed the first genome-wide screen for genes involved in PTM, identifying 30 candidate genes using a cDNA microarray. We have also shown that the genetic architecture underlying the evolution of PTM is dependent upon which time-scale of evolution one is considering. Using next-generation sequencing based SNP markers (RADseq), we constructed comparative Quantitative Trait Locus (QTL) maps that show that the evolution of PTM in response to strong directional selection during post-glacial range expansion of W. smithii has involved different regions of the genome than the evolution of PTM in response to stabilizing selection over longer time-scales.
The evolution of PTM is a complex process, involving many currently unidentified genes. We performed the first genome-wide screen for genes involved in PTM, identifying 30 candidate genes using a cDNA microarray. We have also shown that the genetic architecture underlying the evolution of PTM is dependent upon which time-scale of evolution one is considering. Using next-generation sequencing based SNP markers (RADseq), we constructed comparative Quantitative Trait Locus (QTL) maps that show that the evolution of PTM in response to strong directional selection during post-glacial range expansion of W. smithii has involved different regions of the genome than the evolution of PTM in response to stabilizing selection over longer time-scales.
What is the relationship between the circadian clock controlling daily timing and photoperiodic time measurement controlling seasonal timing?
For over 70 years, most chronobiologists have held that the circadian clock is causally involved in PTM, and for the last decade, the research focus has been on the underlying genetics of this relationship. We showed that the evolution of PTM has been evolutionarily independent of the circadian clock, and established the need to move past a circadian-biased candidate gene approach and towards a forward-genetic, genome-wide approach to the study of photoperiodic timing.
What is the genomic architecture of the earliest stages of population divergence?
In collaboration with Diane Campbell (University of California, Irvine and the Rocky Mountain Biological Laboratory) and Carrie Wu (University of Richmond), we are studying the genomic basis of ecologically mediated reproductive isolation in two sister species of Ipomopsis wildflowers. Preliminary genotype data from the two parental lineages have allowed us to estimate gene flow and to look for signatures of selection. Of ~15,000 loci screened, we are able to find significant divergence at only 10s of loci among these two sibling species – far less than predicted, but consistent with the lack of divergence found in previous studies. We are currently working on determining the candidate functions of genes around divergent loci and working to address the question of why there is such a lack of genetic divergence in two phenotypically divergent species.
What is the genomic architecture of vector competency and Plasmodium immunity in Neotropical malaria vectors?
In collaboration with Anice Sallum (Universidad de Sao Paulo), Jan Conn (Wadsworth Institute), and their students and postdocs, we are studying the population genomics of two Neotropical malaria vectors, Anopheles cruzii, and An. darlingi. We are using genotyping-by-sequencing methods to determine genome-wide patterns of divergence within and among populations.